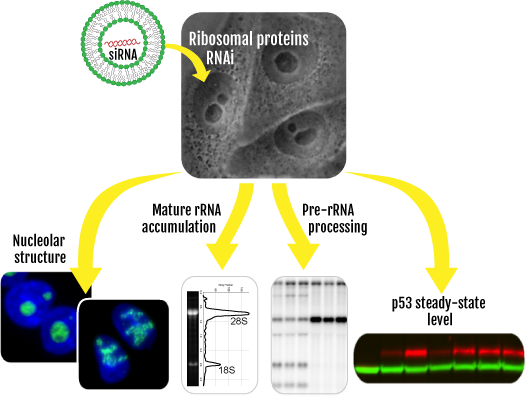
The nucleolus is a dynamic structure whose morphology informs us on the health status of a cell. It is a long-known cancer biomarker and a recently demonstrated therapeutic target. It is therefore important to understand how it is organized structurally and maintained. Ribosomal proteins (r-proteins) control the expression and activity of key tumor suppressors, and they are mutated in multiple cancer predisposition disorders known as ribosomopathies.
Ribosome biogenesis is initiated in the nucleolus where most r-proteins are assembled onto precursor ribosomal RNAs. We systematically depleted all 80 r-proteins in human cells and asked what is the impact on nucleolar structure, mature rRNA accumulation, pre-rRNA processing and p53 steady-state level accumulation.
We developed an unsupervised classification algorithm and defined an index of nucleolar disruption (iNo) to stratify qualitatively and quantitatively nucleolar morphological alterations. We conclude that irrespective of their requirement for processing, the vast majority of r-proteins do not impact nucleolar structure or p53 levels. We found major contributors are all late-assembling large subunit r-proteins. Of these, we identified uL5 and uL18, known to act together with 5S rRNA as an “Hdm2 trap” and p53 stabilizers, or to be part of the central protuberance in mature 60S subunits, as the strongest contributors to nucleolar integrity maintenance.
Our work led us to conclude that the sequence of incorporation of r-proteins into incipient ribosomes is remarkably conserved across evolution, from Bacteria to yeast and man. By identifying the r-proteins that specifically contributes to nucleolar structure maintenance and p53 steady-state accumulation, our work provides novel insights into fundamental aspects of cell and cancer biology.