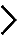Legend : pre-rRNA Processing Legend
3-D models of human ribosomal subunits based on PDB entries 3J3D, 3J3A, 3J3F, and 3J3B. R-proteins are color-coded according to their impact on early, intermediate, or late steps of processing. Left, subunit interface views; right, solvent exposed views. The aminoacyl (A), peptidyl (P) and exit (E) tRNA sites are indicated. Morphological features of the subunits are highlighted. On the LSU: the L1-stalk, the central protuberance (CP) and the phospho-stalk (P-stalk). On the SSU, the beak (Be), head (H), platform (Pt), body (Bd), lef foot (Lf) and right foot (Rf).
Major pre-rRNA intermediates and probes used in this work are schematized between the two heatmaps. Three out of four rRNAs are produced as a long 47S primary transcript by RNA Pol I. The 18S, 5.8S and 28S are separated by noncoding external (ETS) and internal (ITS) transcribed spacers. Probes a, b, and c were used in this work.
From the northern blots all RNA species were quantified with a Phosphorimager, normalized with respect to the nontargeting control (SCR), and their abundances represented as a heatmap using the color code indicated in the middle. The heatmaps were clustered with “R” and grouped in classes of RPs affecting the same processing steps. The different siRNAs used are indicated (#). An asterisk refers to case where the depletion of an RP led to processing defects which belong to two classes. All RNA species were used to cluster the heatmaps but only those directly relevant to synthesis of the small (left part), or large (right part) subunit are shown for simplicity.